Variable Number Tandem Repeats, or VNTRs, are ubiquitous components of the eukaryotic genome, constituting a significant fraction of the non-coding DNA. Understanding the role and function of VNTRs necessitates a grasp of VNTR probes, tools leveraged in molecular biology for their identification and analysis. This exploration delves into the specifics of VNTR probes, their mechanisms, and applications.
What are VNTRs?
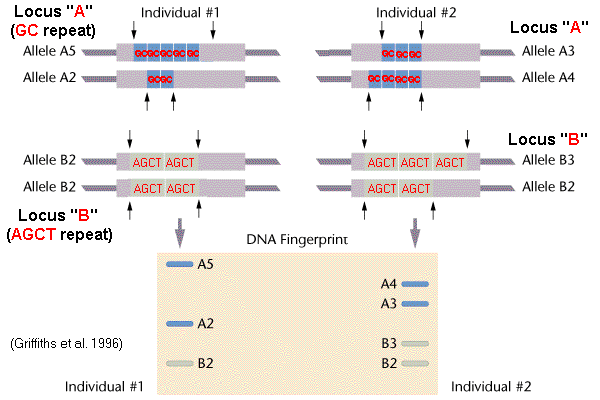
Before discussing probes, it is essential to contextualize VNTRs. These are segments of DNA characterized by short nucleotide sequences that are repeated in tandem at a particular locus on a chromosome. The number of repeats varies widely between individuals, making VNTRs highly polymorphic. This polymorphism is the bedrock of their utility in genetic fingerprinting and population genetics studies. Repeat unit sizes typically range from ten to hundreds of base pairs. Their variable nature, owing to the differing number of repetitions, provides the foundation for their application as molecular markers.
Defining the VNTR Probe
A VNTR probe is a labeled DNA sequence, either single-stranded or double-stranded, complementary to a specific VNTR sequence. This complementarity allows the probe to hybridize, or bind, to the VNTR region within a DNA sample. The labeling, frequently with radioactive isotopes or fluorescent molecules, enables the visualization and detection of the probe-VNTR complex. These probes are meticulously synthesized to target specific VNTR loci, offering researchers a means to pinpoint and analyze these regions within the vast expanse of the genome.
Mechanisms of Action: Hybridization and Detection
The utility of VNTR probes hinges on the principle of nucleic acid hybridization. Here is a step-by-step overview:
- DNA Digestion: Genomic DNA extracted from a sample is digested using restriction enzymes. These enzymes cleave the DNA at specific recognition sites, producing fragments of varying lengths.
- Gel Electrophoresis: The resulting DNA fragments are separated based on their size using gel electrophoresis. Smaller fragments migrate faster through the gel matrix, creating a separation gradient.
- Southern Blotting: The separated DNA fragments are transferred from the gel onto a membrane, typically made of nitrocellulose or nylon. This process, known as Southern blotting, immobilizes the DNA for subsequent probing.
- Probe Hybridization: The membrane is incubated with the labeled VNTR probe. Under appropriate conditions, the probe hybridizes to complementary VNTR sequences present on the membrane. The stringency of hybridization, controlled by factors like temperature and salt concentration, determines the specificity of the probe binding.
- Washing and Detection: After hybridization, the membrane is washed to remove any unbound probe. The bound probe is then detected using a method appropriate for the label employed. Radioactive labels are detected via autoradiography, while fluorescent labels are detected using specialized imaging systems.
Types of VNTR Probes
VNTR probes can be broadly categorized based on their specificity and labeling method:
- Locus-Specific Probes: These probes target a single VNTR locus. They provide information about the allelic variation at that specific location, making them suitable for applications like paternity testing and forensic analysis.
- Multi-Locus Probes: These probes hybridize to multiple VNTR loci scattered throughout the genome. They generate more complex banding patterns, offering a higher degree of discriminatory power, particularly in DNA fingerprinting. However, the complexity of the patterns can make interpretation more challenging.
- Radioactively Labeled Probes: Probes labeled with radioactive isotopes, such as phosphorus-32, were historically prevalent. Autoradiography exposes the radioactive signal emitted, revealing the location of the hybridized probe. While highly sensitive, these probes require stringent safety protocols and specialized equipment.
- Non-Radioactively Labeled Probes: Probes labeled with fluorescent dyes, enzymes, or other non-radioactive tags offer a safer and more convenient alternative. Fluorescent probes are detected using laser scanners or imaging systems, while enzyme-linked probes generate a colored product that can be visualized directly.
Applications of VNTR Probes
VNTR probes have found wide-ranging applications across diverse fields:
- DNA Fingerprinting: VNTR probes revolutionized forensic science. The unique VNTR profiles of individuals can be used to establish identity or exclude suspects in criminal investigations. The probability of two unrelated individuals having identical VNTR profiles is extremely low, providing strong evidence for identification.
- Paternity Testing: VNTR analysis can definitively establish parentage. A child inherits VNTR alleles from both parents, and the VNTR profile of a child must be consistent with the profiles of the alleged parents. Discrepancies between the profiles can exclude a potential parent with a high degree of certainty.
- Population Genetics: VNTR probes are invaluable tools for studying genetic diversity and population structure. By analyzing the frequencies of different VNTR alleles in various populations, researchers can infer patterns of migration, ancestry, and genetic relatedness.
- Disease Diagnosis: While not as widely used as in the past due to the rise of other techniques, VNTRs located near or within genes associated with certain diseases can be used as markers for disease susceptibility. Changes in VNTR repeat number can sometimes be directly linked to disease phenotypes.
- Plant and Animal Breeding: VNTR probes are used in agriculture to identify desirable traits in livestock and crops. They can assist in marker-assisted selection, where individuals with specific VNTR alleles associated with desirable traits are selectively bred. This accelerates the breeding process and improves the efficiency of crop and livestock improvement.
Advantages and Limitations
VNTR probes offer several advantages, including high discriminatory power, relative ease of use, and established protocols. However, they also have limitations. Southern blotting, the traditional method for VNTR analysis, is time-consuming and requires relatively large amounts of DNA. The interpretation of multi-locus VNTR profiles can be complex. Moreover, the use of radioactive probes raises safety concerns.
The Shift Towards STRs and Alternative Technologies
While VNTR probes were a cornerstone of molecular biology for many years, they have gradually been superseded by the use of Short Tandem Repeats (STRs), also known as microsatellites. STRs are shorter than VNTRs, making them more amenable to analysis using Polymerase Chain Reaction (PCR) based methods. PCR allows for the amplification of specific DNA regions, requiring only minute amounts of starting material and dramatically accelerating the analysis process. Multiplex PCR, where multiple STR loci are amplified simultaneously, has further streamlined DNA fingerprinting and related applications. Next-generation sequencing (NGS) technologies also provide alternative means of analyzing VNTR regions, offering even higher throughput and resolution.
Despite the rise of these newer technologies, understanding VNTR probes provides valuable insight into the historical development of molecular biology techniques and underscores the principles of nucleic acid hybridization and polymorphism analysis. While their prominence in routine applications may have diminished, the foundational knowledge derived from their use remains relevant and continues to inform contemporary genomic research.








Leave a Comment