In the realm of forensic science and genetic analysis, the terms DNA profiling and DNA fingerprinting often surface, frequently used interchangeably. But are they truly synonymous? The answer, while seemingly simple, requires a nuanced understanding of their historical context, technological evolution, and subtle differences in application.
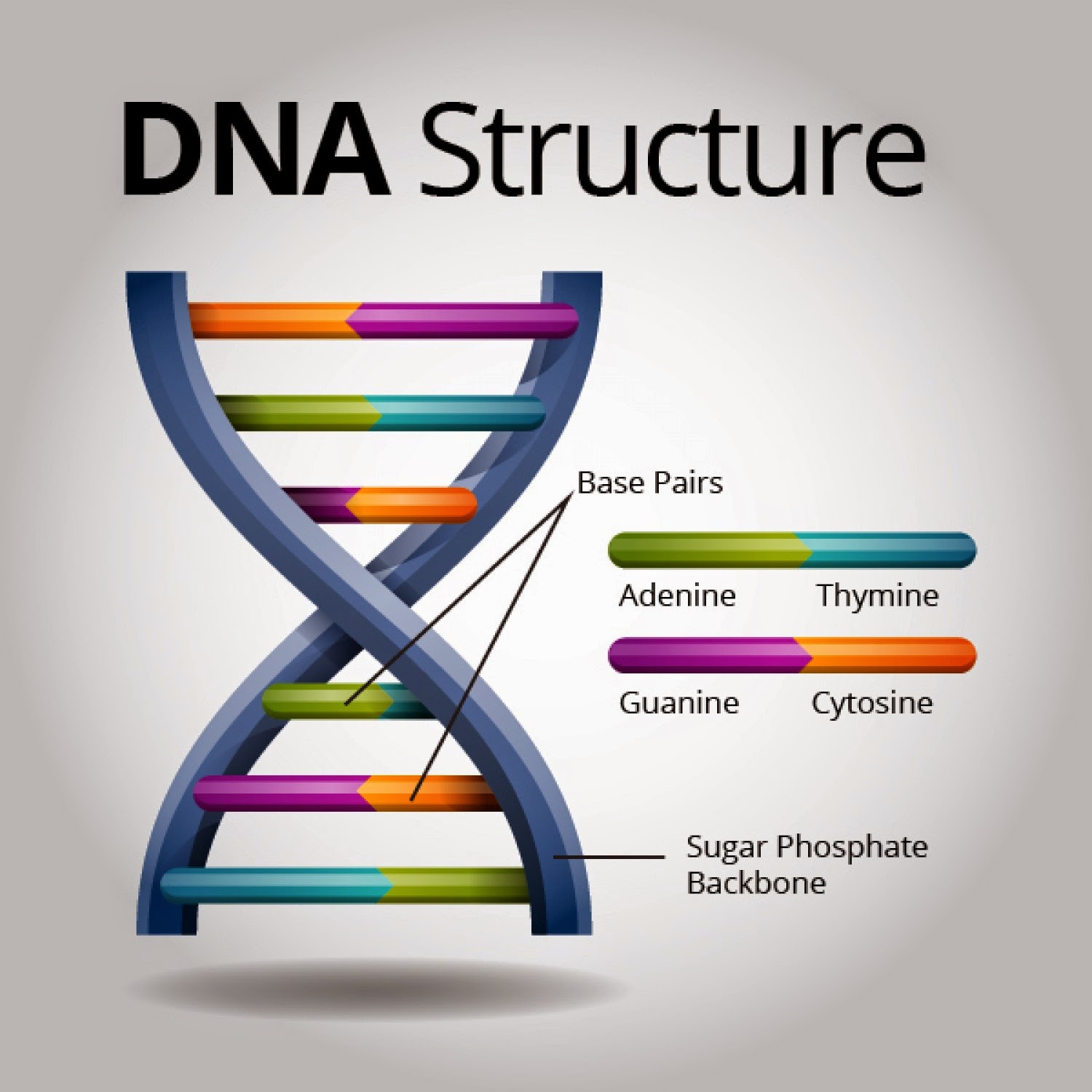
At their core, both DNA profiling and DNA fingerprinting aim to achieve the same goal: to create a unique identifier for an individual based on their deoxyribonucleic acid (DNA). Think of DNA as the intricate blueprint of life, a complex code residing within every cell, dictating everything from eye color to susceptibility to certain diseases. The beauty, and utility, lies in the fact that, save for identical twins, no two individuals share the exact same DNA sequence. This inherent variability is what both techniques exploit.
A Historical Perspective: From Fingerprints to Genetic Signatures
The term DNA fingerprinting predates DNA profiling. It emerged in the mid-1980s, pioneered by British geneticist Sir Alec Jeffreys. His groundbreaking work involved analyzing highly variable regions of DNA known as Variable Number Tandem Repeats (VNTRs). These VNTRs are segments of DNA where a short nucleotide sequence is repeated a variable number of times. The length of these repeated sequences differs considerably between individuals, making them ideal markers for identification.
Jeffreys’ original technique was akin to creating a bar code, or more precisely, a unique series of bands on a gel. This “fingerprint” was generated by digesting DNA with restriction enzymes, separating the resulting fragments by size using gel electrophoresis, and then probing for specific VNTR sequences. The resulting pattern of bands was then compared to determine if two samples originated from the same individual. It was revolutionary, ushering in a new era of forensic science and paternity testing. The initial application was indeed evocative of traditional fingerprinting, hence the name.
DNA profiling, on the other hand, represents a more modern and refined approach. It leverages different genetic markers known as Short Tandem Repeats (STRs). STRs, as the name implies, are shorter than VNTRs and are easier to amplify using Polymerase Chain Reaction (PCR). PCR is a molecular biology technique that allows for the exponential amplification of a specific DNA sequence, even if only a minute amount of starting material is available. This is a significant advantage, as forensic samples are often degraded or available in limited quantities.
The Rise of STR Analysis: A Paradigm Shift
The shift from VNTR to STR analysis was driven by several factors. First, STR analysis is more amenable to automation, allowing for high-throughput processing of samples. Second, the smaller size of STRs makes them less susceptible to degradation, a critical consideration in forensic casework. Third, standardized STR marker sets have been developed, enabling the creation of large DNA databases, such as CODIS (Combined DNA Index System) in the United States. These databases allow for the efficient comparison of DNA profiles across different jurisdictions, significantly enhancing law enforcement capabilities.
Think of STR analysis as creating a unique numerical code rather than a visual bar code. Each STR locus (location on a chromosome) is assigned a number based on the number of repeats it contains. A DNA profile consists of a series of these numbers, one for each STR locus analyzed. This numerical format facilitates computerized analysis and comparison, making it far more efficient than the manual interpretation required for VNTR-based fingerprints.
Subtle Differences, Significant Implications
While the terms are often used interchangeably, it is crucial to recognize that DNA profiling, using STR analysis, has largely superseded DNA fingerprinting, which relies on VNTR analysis. DNA profiling is the current standard in forensic science, paternity testing, and other applications requiring individual identification.
The difference extends beyond just the markers used. The statistical analysis employed to interpret the results also differs. With VNTR analysis, the calculation of match probabilities was more complex due to the continuous nature of VNTR allele sizes. With STR analysis, the discrete nature of allele sizes (whole number repeats) simplifies the statistical calculations and allows for more precise estimation of the rarity of a particular DNA profile.
Consider this analogy: DNA fingerprinting is like using a broad brush to paint a picture, capturing the general outline and characteristics. DNA profiling, on the other hand, is like using a fine-tipped pen to add intricate details, resulting in a more precise and informative representation.
The Broader Context: Applications Beyond Forensics
Both DNA fingerprinting (historically) and DNA profiling (currently) have applications extending far beyond the realm of criminal justice. They are used in:
- Paternity testing: To definitively establish biological parentage.
- Medical diagnostics: To identify genetic predispositions to certain diseases.
- Anthropology: To study human migration patterns and population genetics.
- Wildlife conservation: To track endangered species and combat poaching.
- Agriculture: To identify and protect valuable plant varieties.
In Conclusion: A Question of Precision and Modernity
In essence, while both DNA profiling and DNA fingerprinting serve the purpose of individual identification based on DNA, they represent distinct methodologies. DNA fingerprinting is the historical precursor, while DNA profiling is the modern, refined, and standardized technique. The shift from VNTRs to STRs marked a significant advancement, improving the efficiency, accuracy, and applicability of DNA analysis. Using the term DNA fingerprinting to describe modern STR-based profiling, while common, lacks the technical precision demanded by contemporary scientific discourse. DNA profiling is the method used in modern analysis.








Leave a Comment