DNA fingerprinting, a cornerstone of modern forensic science and genetic research, hinges on the remarkable premise that each individual possesses a unique genomic signature. This signature isn’t etched in immutable code across the entire genome, but rather resides within specific, highly variable regions. Understanding what constitutes these differences is paramount to appreciating the power and limitations of this ubiquitous technology.
At its core, DNA fingerprinting involves identifying differences in highly repetitive sequences of DNA. Imagine your genome as a vast library filled with books, most of which contain the same core information necessary for cellular function. However, interspersed throughout this library are numerous short stories, copied and pasted multiple times, with slight variations in their repetition and arrangement. These “short stories” are the highly repetitive sequences that DNA fingerprinting exploits.
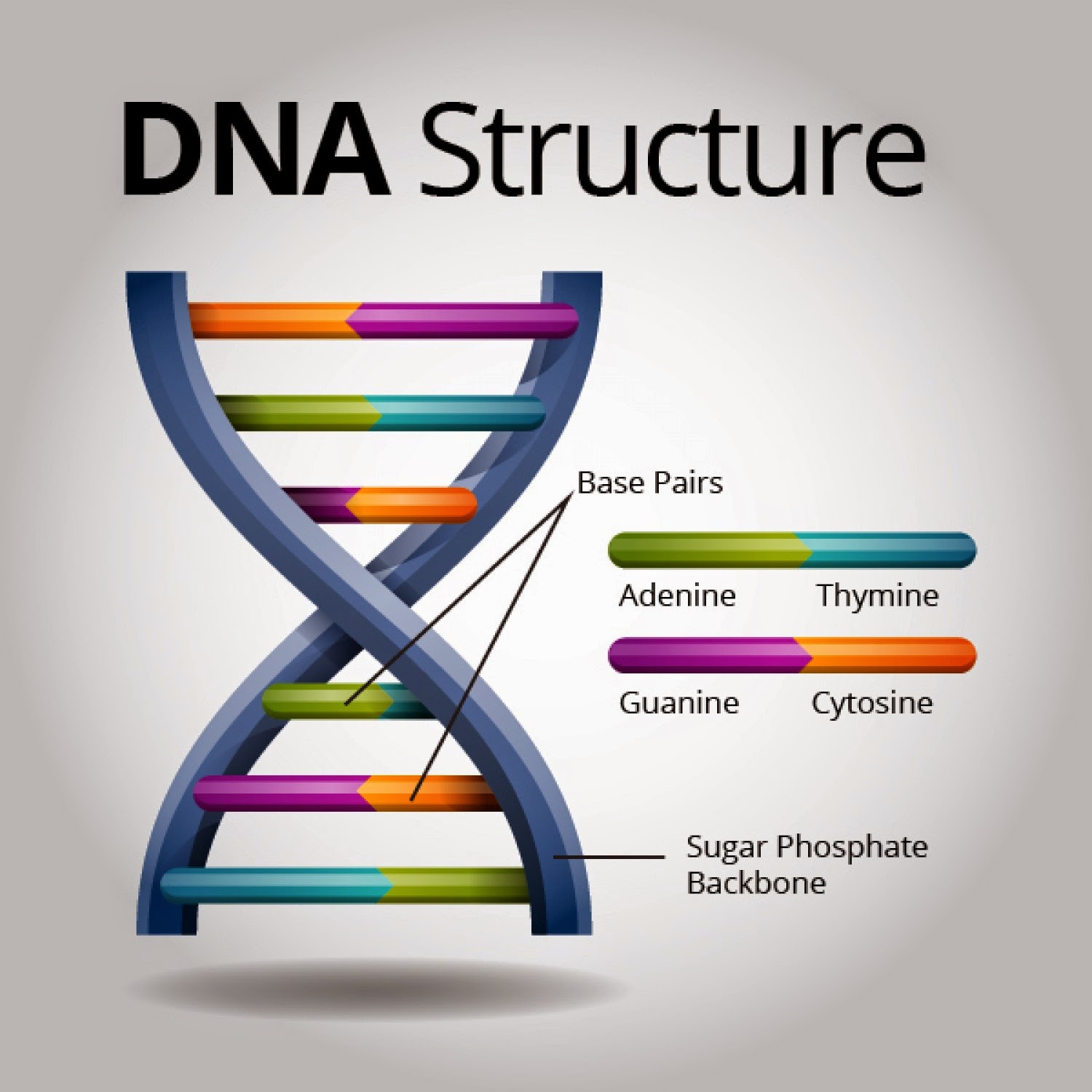
More precisely, these regions consist of Variable Number Tandem Repeats (VNTRs) and Short Tandem Repeats (STRs). VNTRs, as their name implies, are sequences of DNA, typically 10-100 base pairs long, that are repeated in tandem (one after another) at specific locations, or loci, within the genome. The number of times a particular VNTR sequence is repeated varies significantly between individuals. One person might have 7 repeats at a specific locus, while another might have 12, and a third might have only 3. This variation in repeat number is the key to distinguishing individuals.
STRs, also known as microsatellites, are similar to VNTRs but involve much shorter repeat units, usually 2-6 base pairs in length. The smaller size of STRs makes them more amenable to analysis using polymerase chain reaction (PCR), a technique that amplifies specific DNA sequences. PCR allows scientists to generate millions of copies of the STR regions, even from tiny samples of DNA, such as those found at crime scenes. This amplification process is crucial, as it provides sufficient material for accurate analysis.
The power of DNA fingerprinting doesn’t solely rely on analyzing a single VNTR or STR locus. Instead, forensic scientists and researchers typically examine multiple loci simultaneously. Consider this analogy: trying to identify a single person based on the color of their hair might lead to numerous false matches. However, if you combine hair color with eye color, height, and other distinguishing features, the likelihood of a unique identification increases dramatically. Similarly, by analyzing multiple VNTR or STR loci, the probability of two unrelated individuals having the same DNA profile becomes infinitesimally small.
Furthermore, DNA fingerprinting discerns differences in allele sizes at these repetitive loci. An allele is simply a variant form of a gene or DNA sequence. In the context of VNTRs and STRs, different repeat numbers represent different alleles. During DNA fingerprinting, the lengths of the amplified DNA fragments containing the VNTRs or STRs are measured. These length differences, corresponding to different allele sizes, are what allow for individual identification.
It’s crucial to understand that DNA fingerprinting generally does not analyze coding regions of the genome, the regions that contain the instructions for making proteins. Instead, it focuses on the non-coding, repetitive sequences that are less susceptible to evolutionary pressures. Coding regions are highly conserved, meaning they tend to be similar across individuals, as mutations in these regions can have detrimental effects on cellular function. Non-coding regions, on the other hand, are more tolerant of variations, making them ideal for distinguishing individuals without affecting essential biological processes.
The technique also unveils differences related to fragment length polymorphisms. Polymorphism simply means “many forms.” Fragment length polymorphism refers to the variation in the length of DNA fragments generated by cutting the DNA with restriction enzymes. Restriction enzymes are molecular scissors that recognize and cut DNA at specific sequences. The length of the resulting fragments will vary depending on the presence or absence of these recognition sites within the VNTR or STR regions. This variation in fragment length is then visualized using gel electrophoresis, a technique that separates DNA fragments based on their size. Shorter fragments migrate faster through the gel than longer fragments, creating a pattern of bands that represents the individual’s DNA fingerprint.
In essence, DNA fingerprinting leverages the seemingly chaotic and redundant aspects of the genome – the repetitive sequences – to establish a unique identifier. It is a testament to how seemingly unimportant regions of our genetic code can hold the key to individual identity. Think of it as finding order within chaos, creating a personalized barcode from the genetic noise that surrounds the functional genes. The technology has revolutionized fields ranging from criminal justice to paternity testing and population genetics. It enables us to unravel complex mysteries, connect individuals to their past, and understand the intricacies of genetic diversity. The seemingly simple process of identifying differences in repetitive DNA sequences has unlocked a Pandora’s Box of possibilities, transforming our understanding of life itself.








Leave a Comment