Polymerase Chain Reaction (PCR) is a ubiquitous technique in molecular biology, employed to amplify specific DNA sequences. While the amplification process itself is crucial, the subsequent analysis of the resultant PCR products is equally important for verifying the success of the reaction and gleaning meaningful insights. Analyzing PCR products involves a panoply of techniques, each offering unique advantages and suited for different applications. Let’s delve into some of the most common and effective methods for analyzing PCR amplicons.
Gel Electrophoresis: A Foundational Technique
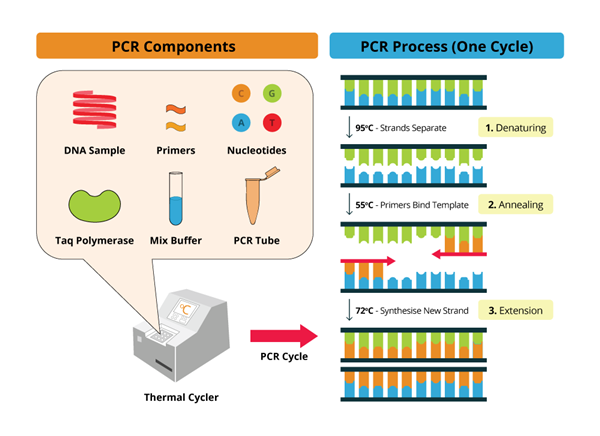
Gel electrophoresis remains a cornerstone method for visualizing and assessing PCR products. This technique separates DNA fragments based on their size and charge, facilitating a qualitative assessment of the amplification. Typically, PCR products are loaded into wells of an agarose or polyacrylamide gel, and an electric field is applied. DNA, being negatively charged due to its phosphate backbone, migrates towards the positive electrode (anode). Smaller fragments navigate the gel matrix more readily, resulting in size-dependent separation.
Following electrophoresis, the gel is stained with a DNA-binding dye, such as ethidium bromide or SYBR Green, which intercalates between DNA base pairs and fluoresces under ultraviolet (UV) light. This allows for visualization of the DNA bands, each representing a population of DNA fragments of a specific size. The presence of a band at the expected size confirms the amplification of the target sequence. The intensity of the band can provide a semi-quantitative estimate of the amount of PCR product generated.
Furthermore, gel electrophoresis can reveal non-specific amplification, indicated by the presence of bands at unexpected sizes. It can also detect primer dimers, which are short, undesired products formed by the self-annealing of primers. Troubleshooting PCR reactions often begins with a meticulous examination of the gel to identify and mitigate these issues.
Restriction Enzyme Digestion: Validating Sequence Specificity
Restriction enzyme digestion offers a more rigorous approach to verifying the identity of PCR products. Restriction enzymes, also known as restriction endonucleases, are enzymes that cleave DNA at specific recognition sequences. By designing primers that introduce restriction enzyme sites flanking the amplified region, the PCR product can be subjected to digestion with the corresponding enzyme.
The resulting fragments are then analyzed by gel electrophoresis. If the PCR product contains the expected restriction enzyme site, digestion will produce fragments of predictable sizes. Comparing the observed fragment sizes with the expected sizes confirms the presence of the intended sequence. This technique is particularly useful for confirming the insertion of a specific DNA fragment into a vector or for detecting single nucleotide polymorphisms (SNPs) that alter restriction enzyme recognition sites.
Southern Blotting: Detecting Specific Sequences in Complex Mixtures
Southern blotting, named after its inventor Edwin Southern, is a technique used to detect specific DNA sequences within a complex mixture of DNA fragments. This method involves separating DNA fragments by gel electrophoresis, transferring them to a membrane (typically nitrocellulose or nylon), and then hybridizing the membrane with a labeled DNA probe complementary to the target sequence.
The probe is labeled with a radioactive isotope or a non-radioactive reporter molecule, allowing for its detection after hybridization. Following washing to remove unbound probe, the membrane is exposed to X-ray film (for radioactive probes) or treated with reagents to reveal the location of the bound probe (for non-radioactive probes). This technique is particularly valuable for detecting rare or low-abundance PCR products within a complex sample, such as genomic DNA.
DNA Sequencing: The Gold Standard for Verification
DNA sequencing represents the gold standard for confirming the identity and integrity of PCR products. Sequencing determines the precise nucleotide sequence of the amplified DNA, providing unambiguous confirmation of its identity. Sanger sequencing, also known as chain-termination sequencing, is a widely used method for sequencing PCR products. In this technique, DNA polymerase is used to synthesize a complementary strand of the PCR product in the presence of dideoxynucleotides (ddNTPs), which terminate DNA synthesis.
Each ddNTP is labeled with a different fluorescent dye, allowing for the identification of the terminating nucleotide at each position. The resulting fragments are separated by capillary electrophoresis, and the sequence is determined based on the order of the fluorescent dyes. Next-generation sequencing (NGS) technologies offer higher throughput and are particularly useful for analyzing complex mixtures of PCR products or for sequencing long DNA fragments. Sequencing is essential for validating the accuracy of PCR amplification and for identifying any mutations or sequence variations.
Quantitative PCR (qPCR): Measuring Amplicon Abundance
Quantitative PCR (qPCR), also known as real-time PCR, is a technique used to measure the amount of PCR product generated during the amplification process. This technique allows for the quantification of the starting amount of target DNA in a sample. qPCR relies on the use of fluorescent dyes or probes that bind to the PCR product and emit fluorescence as the amount of product increases. The fluorescence signal is measured in real-time, allowing for the determination of the cycle threshold (Ct) value, which is the number of cycles required for the fluorescence signal to reach a predetermined threshold.
The Ct value is inversely proportional to the starting amount of target DNA. By comparing the Ct values of unknown samples with those of known standards, the amount of target DNA in the unknown samples can be quantified. qPCR is widely used for gene expression analysis, pathogen detection, and other applications where accurate quantification of DNA is required.
High-Resolution Melting (HRM) Analysis: Detecting Sequence Variations
High-resolution melting (HRM) analysis is a post-PCR technique used to detect sequence variations, such as SNPs and insertions/deletions, in PCR products. This technique involves heating the PCR product in the presence of a saturating DNA dye and monitoring the fluorescence signal as the DNA melts. The melting temperature of DNA is dependent on its sequence, with different sequences exhibiting different melting profiles.
By comparing the melting profiles of different PCR products, sequence variations can be detected. HRM analysis is a rapid and cost-effective method for screening for sequence variations in a large number of samples. It is often used for genotyping, mutation scanning, and species identification.
In conclusion, the analysis of PCR products is a crucial step in ensuring the validity and utility of PCR-based experiments. From the foundational technique of gel electrophoresis to the precision of DNA sequencing and the quantitative power of qPCR, a diverse toolkit exists to scrutinize amplicons. The selection of an appropriate analysis method hinges on the specific research question, the desired level of detail, and the available resources. As PCR continues to be a dominant force in molecular biology, a thorough understanding of these analytical techniques is paramount for extracting maximum value from this powerful technology.








Leave a Comment