Ever pondered the molecular equivalent of a clumsy key jammed in the wrong lock? The world of restriction enzymes, those diligent molecular scissors, presents precisely that analogy. Employing the incorrect restriction enzyme can instigate a cascade of deleterious consequences, impacting everything from simple cloning experiments to intricate genomic analyses. Let’s delve into why fidelity in restriction enzyme selection is paramount.
I. The Specificity Paradigm: Recognizing Palindromic Sequences
Restriction enzymes, or restriction endonucleases, are not indiscriminate cleavers. Their functionality hinges on recognizing and binding to specific DNA sequences, often palindromic. Palindromic sequences, in this context, are nucleotide sequences that read the same forwards and backward on complementary strands. Consider the EcoRI enzyme, which meticulously targets the GAATTC sequence. Introduce EcoRI to a DNA strand devoid of this sequence, and it will stand idly by, incapable of executing its scission. Conversely, if a different restriction enzyme with a different specificity, such as BamHI (GGATCC), is used it will only cleave if that specific sequence is available. This specificity is not merely a biochemical quirk; it forms the bedrock of predictable and controlled DNA manipulation. Incorrectly selecting an enzyme leads to inefficient cutting, or worse, cutting at unintended sites if any.
II. The Perils of Off-Target Cleavage: Aberrant Fragmentation
What happens when the wrong restriction enzyme finds its way into the experimental arena? The outcome isn’t merely a lack of digestion; it’s the potential for off-target cleavage. While restriction enzymes are designed to be highly specific, under suboptimal conditions, such as high enzyme concentration, non-ideal buffer compositions, or extended incubation times, some enzymes exhibit relaxed specificity. This phenomenon, sometimes referred to as “star activity,” involves the enzyme cleaving sequences that are similar, but not identical, to its cognate recognition site. The consequence? Unforeseen DNA fragmentation, rendering subsequent cloning or analysis exercises futile. Aberrant fragmentation undermines the integrity of the DNA construct, leading to unpredictable results and wasted resources. Therefore, it is important to carefully match the enzyme to the specific sequence available in the desired location.
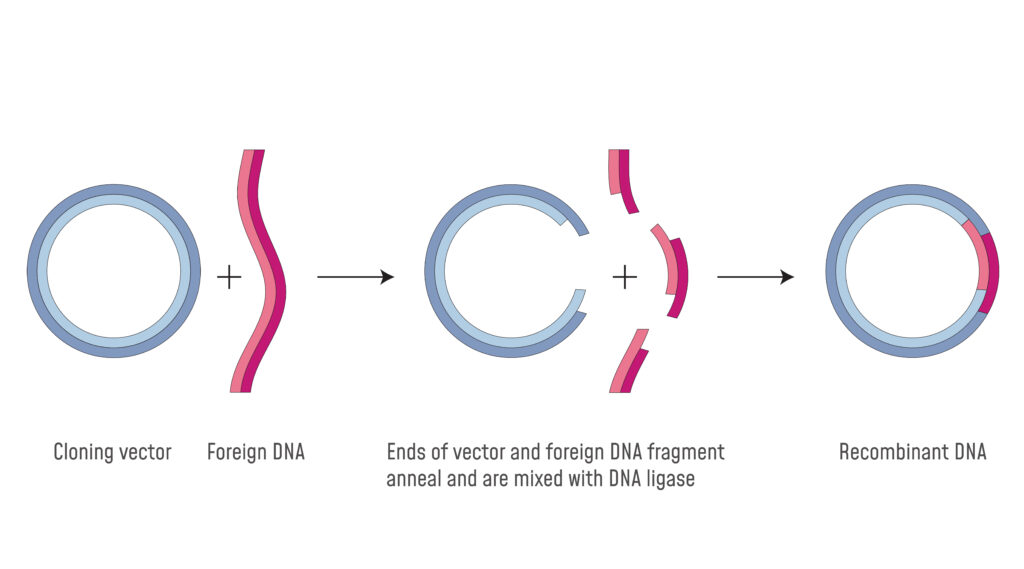
III. Compromised Ligation Efficiency: Fragment Incompatibility
Restriction enzymes don’t simply cleave; they dictate the nature of the DNA ends they generate. These ends can be either “sticky” (possessing single-stranded overhangs) or “blunt” (flush ends). Sticky ends, such as those produced by EcoRI, are particularly conducive to ligation, as their overhangs facilitate complementary base pairing with other fragments generated by the same enzyme. Blunt ends, while versatile, require higher concentrations of ligase for efficient joining. When incompatible ends arise from inappropriate restriction enzyme selection, the ligation process becomes severely hampered. Imagine attempting to ligate a fragment with EcoRI-generated overhangs to a fragment with blunt ends – the union is highly improbable, if not impossible. This inefficiency translates into reduced cloning efficiency, prolonged experimental timelines, and increased frustration.
IV. Disrupted Reading Frames: Mutations and Non-Functional Proteins
In the realm of recombinant protein expression, the stakes are even higher. Incorrect restriction enzyme selection can wreak havoc on the open reading frame (ORF) of a gene. The ORF, the contiguous stretch of DNA that encodes a protein, is exquisitely sensitive to insertions and deletions. If restriction sites are improperly placed, or if unintended cleavage occurs within the ORF, the reading frame can be shifted. A frameshift mutation alters the codon sequence downstream of the insertion or deletion, leading to the production of a non-functional or truncated protein. This can manifest as a complete loss of protein activity, or, in some cases, the expression of a protein with deleterious properties. In essence, a single incorrect restriction enzyme choice can negate the entire purpose of a gene cloning and expression endeavor.
V. Impediments to Diagnostic Accuracy: False Positives and Negatives
Restriction enzymes play a pivotal role in various diagnostic techniques, such as restriction fragment length polymorphism (RFLP) analysis. RFLP analysis exploits the natural variation in DNA sequences between individuals to identify genetic markers. By digesting DNA with specific restriction enzymes and analyzing the resulting fragment patterns, researchers can detect subtle differences in DNA sequence that correlate with disease susceptibility or other traits. However, the use of an incorrect restriction enzyme can compromise the accuracy of RFLP analysis. If the chosen enzyme fails to cleave at a critical site, or if it cleaves at an unintended site, the resulting fragment pattern will be distorted, leading to false positives or false negatives in the diagnostic assessment. This can have serious implications for clinical decision-making.
VI. Compromising DNA Mapping and Sequencing: Erroneous Data
Restriction enzymes are indispensable tools in DNA mapping and sequencing projects. Restriction mapping involves generating a “fingerprint” of a DNA molecule by digesting it with a panel of restriction enzymes and analyzing the resulting fragment sizes. This information can be used to create a physical map of the DNA, indicating the relative positions of various restriction sites. Similarly, restriction enzymes are often used to generate DNA fragments of appropriate sizes for sequencing. If an incorrect enzyme is selected, the resulting fragment patterns will be inaccurate, leading to an erroneous DNA map. In sequencing applications, incorrect enzyme choices can lead to incomplete or overlapping data, making it difficult to assemble a contiguous sequence. The domino effect leads to inaccurate data.
VII. The Economic Repercussions: Wasted Time and Resources
Beyond the scientific ramifications, using the wrong restriction enzyme carries significant economic consequences. Research endeavors are costly undertakings, demanding substantial investments in reagents, equipment, and personnel time. Incorrect enzyme selection can render entire experiments invalid, necessitating repetition and consuming valuable resources. The cumulative effect of multiple such errors can quickly escalate, leading to significant budgetary overruns and project delays. Careful planning, meticulous attention to detail, and a thorough understanding of enzyme specificity are essential to mitigate these economic risks.
In summation, the seemingly simple act of choosing a restriction enzyme is fraught with potential pitfalls. Selecting the appropriate enzyme is not merely a matter of convenience; it is a fundamental prerequisite for ensuring the accuracy, efficiency, and ultimately, the success of molecular biology experiments. The consequences of error range from inefficient ligation to disrupted reading frames, compromised diagnostic accuracy, and significant economic losses. By embracing diligence, precision, and a deep understanding of enzyme specificity, researchers can navigate the complex world of restriction enzymes with confidence and achieve their scientific objectives.








Leave a Comment