Restriction enzymes, also known as restriction endonucleases, are ubiquitous molecular scalpels in the realm of molecular biology. These enzymes are bacterial proteins that cleave double-stranded DNA at specific sequences known as recognition sites. Think of them as highly precise scissors, cutting DNA only when they encounter a specific genetic code.
The Role of Restriction Enzymes in Bacteria
In bacteria, restriction enzymes serve as a crucial defense mechanism against foreign DNA, such as that from viruses (bacteriophages). When a bacteriophage infects a bacterium, it injects its DNA into the cell. The bacterium’s restriction enzymes can then recognize and cleave this foreign DNA, effectively neutralizing the viral threat. This process is called restriction. To protect their own DNA from being cleaved by their own restriction enzymes, bacteria use a companion enzyme called a methylase. Methylases modify the bacterial DNA at or near the restriction enzyme recognition site, thus preventing the restriction enzyme from binding and cutting.
Nomenclature of Restriction Enzymes
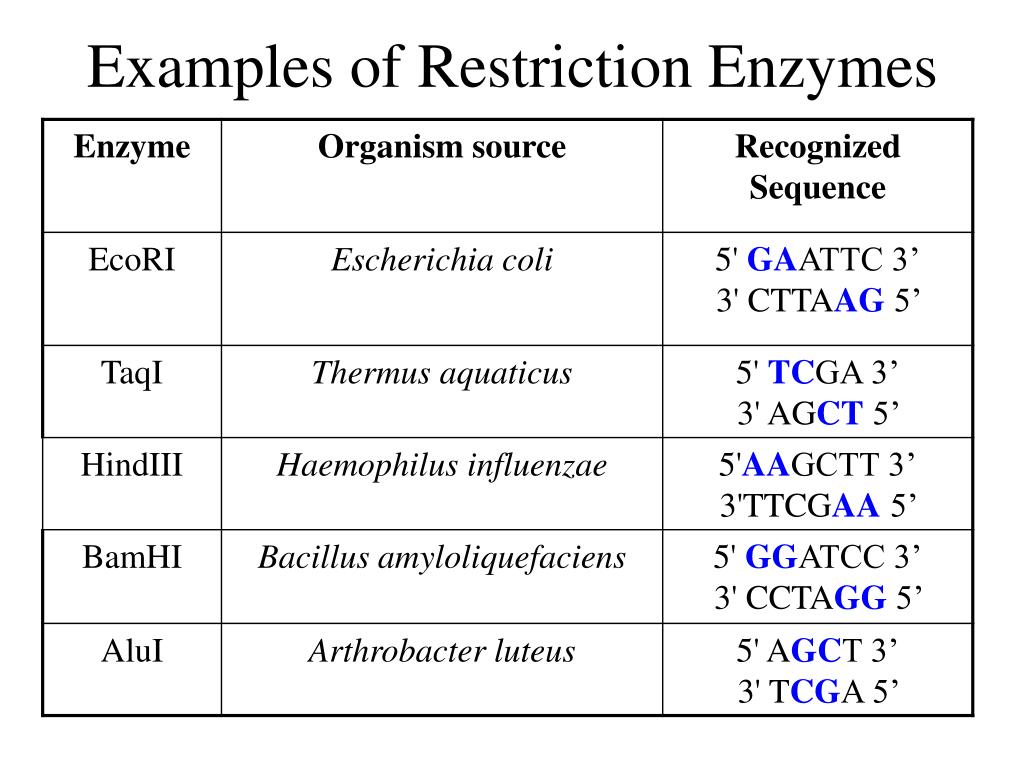
The naming convention for restriction enzymes is quite systematic, lending clarity to their identification. Each enzyme’s name is derived from the bacterial species it originates from. For instance, EcoRI comes from Escherichia coli RY13. The first letter denotes the genus (E), the next two letters represent the species (co), and the following letter indicates the strain (R). The Roman numeral indicates the order in which the enzyme was discovered and isolated from that particular strain. This standardized nomenclature helps to avoid confusion and ensures clarity in scientific communication. A newly discovered restriction enzyme from a new bacterial species would be named according to the above stated pattern.
Types of Restriction Enzymes
Restriction enzymes are categorized into four main types (Type I, II, III, and IV), based on their structure, cofactor requirements, recognition sites, and cleavage mechanisms. The majority of restriction enzymes used in molecular biology laboratories are Type II. Type II restriction enzymes are highly valued for their simplicity and predictable cleavage patterns.
Type I enzymes are complex, multi-subunit proteins that bind to a specific recognition sequence but cleave the DNA at a random site far from the recognition sequence. They require ATP, Mg2+, and S-adenosylmethionine (SAM) for their activity.
Type II enzymes, in contrast, are simpler and cleave DNA within or at specific distances from the recognition sequence. They typically require only Mg2+ for activity. Type II restriction enzymes are the most commonly used in laboratory settings due to their predictability.
Type III enzymes are also complex and require ATP and Mg2+ for activity. They recognize specific sequences but cleave the DNA at a defined distance from the recognition site, albeit less precisely than Type II enzymes.
Type IV enzymes target modified DNA, such as methylated, hydroxymethylated, or glucosyl-hydroxymethylated DNA. Their recognition sites are not well defined, and their cleavage mechanisms are still under investigation.
Recognition Sites and Cleavage Patterns
Restriction enzymes recognize specific DNA sequences, usually 4 to 8 base pairs in length. These sequences are often palindromic, meaning that they read the same forward on one strand as they do backward on the complementary strand. The cleavage pattern can result in either “sticky ends” or “blunt ends.”
Sticky ends are staggered cuts that leave single-stranded overhangs. These overhangs can base-pair with complementary sticky ends generated by the same restriction enzyme, facilitating the joining of DNA fragments. Blunt ends, on the other hand, are cuts that create flush ends without any overhangs. Blunt-ended fragments can be ligated to any other blunt-ended fragment, but the ligation is less efficient than with sticky ends.
Example 1: EcoRI
EcoRI, derived from Escherichia coli, is a classic example of a Type II restriction enzyme. Its recognition sequence is 5′-GAATTC-3′, and it cleaves the DNA between the G and A bases, creating sticky ends. The resulting overhang is 5′-AATT-3′. EcoRI is frequently used in molecular cloning to insert DNA fragments into plasmids.
Imagine you have a circular plasmid DNA and a linear piece of DNA that you want to insert into the plasmid. Both the plasmid and the DNA fragment are digested with EcoRI. The enzyme cleaves the plasmid at a single EcoRI site, linearizing it and creating 5′-AATT-3′ overhangs. The DNA fragment is also cleaved with EcoRI, generating compatible 5′-AATT-3′ overhangs. When these two DNA fragments are mixed together, the complementary sticky ends anneal, bringing the DNA fragment and the plasmid together. DNA ligase is then used to seal the phosphodiester bonds, creating a recombinant plasmid containing the inserted DNA fragment. This process is fundamental to genetic engineering.
Example 2: HindIII
HindIII, isolated from Haemophilus influenzae, is another commonly used Type II restriction enzyme. It recognizes the sequence 5′-AAGCTT-3′ and cleaves between the A bases, creating sticky ends with a 5′-AGCT-3′ overhang. Like EcoRI, HindIII is widely employed in recombinant DNA technology.
Consider a situation where you need to cut a specific gene out of a larger DNA molecule. You could design PCR primers that flank the gene of interest and incorporate HindIII restriction sites at their 5′ ends. After PCR amplification, the resulting PCR product will have HindIII sites at both ends. Digesting the PCR product with HindIII will generate sticky ends, allowing you to clone the gene into a plasmid that has also been digested with HindIII. This strategy allows for precise manipulation of DNA sequences and is a cornerstone of modern molecular biology.
Applications of Restriction Enzymes
Restriction enzymes are indispensable tools in molecular biology, finding applications in a wide array of techniques, including:
- DNA Cloning: As demonstrated above, restriction enzymes are essential for creating recombinant DNA molecules.
- DNA Fingerprinting: Restriction fragment length polymorphism (RFLP) analysis utilizes restriction enzymes to identify variations in DNA sequences.
- Gene Mapping: Restriction enzymes are used to create physical maps of genomes.
- Southern Blotting: Restriction enzymes are used to digest DNA prior to hybridization with a probe in Southern blotting.
- Site-Directed Mutagenesis: Restriction enzymes can be used to introduce specific mutations into DNA.
- Diagnostics: Restriction enzymes can be used to detect specific DNA sequences in diagnostic assays.
In conclusion, restriction enzymes are invaluable tools in molecular biology, enabling scientists to manipulate and analyze DNA with precision and control. Their discovery and characterization have revolutionized the field, paving the way for advancements in genetic engineering, biotechnology, and medicine.








Leave a Comment